1
Microbial Genetics
The science of genetics defines and analyzes heredity of the vast array of
structural and physiologic functions that form the properties of organisms.
The basic unit of heredity is the gene, a segment of deoxyribonucleic acid (DNA)
that encodes in its nucleotide sequence information for a specific physiologic
property.
DNA as the fundamental element of heredity was suggested in the 1930s
from a seminal experiment performed by Frederick Griffith. MacLeod, and
McCarty discovered that DNA was the transforming agent.
Nucleic acid Structure
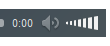
Genetic information in bacteria is stored as a sequence of DNA bases, The
orientation of the two DNA strands is antiparallel. One strand is chemically
oriented in a 5′→3′ direction, and its complementary strand runs 3′→5′. The
complementarity of the bases enables one strand (template strand) to provide the
information for copying or expression of information in the other strand (coding
strand).
Microbiology
Medical bacteriology
Dr. Zainab D. Degaim

2
Nucleic acids are large polymers consisting of repeating nucleotide units.
Each nucleotide contains one phosphate group, one pentose or
deoxypentose sugar, and one purine or pyrimidine base.
In DNA the sugar is D-2-deoxyribose; in RNA the sugar is D-ribose.
In DNA the purine bases are adenine (A) and guanine (G), and the
pyrimidine bases are thymine (T) and cytosine (C).
In RNA, uracil (U) replaces thymine.
The repeating structure of polynucleotides involves alternating sugar and
phosphate residues, with phosphodiester bonds linking the 3'-hydroxyl
group of one nucleotide sugar to the 5'- hydroxyl group of the adjacent
nucleotide sugar. A purine or pyrimidine base is linked at the 1'-carbon
atom of each sugar residue and projects from the repeating sugarphosphate
backbone.
Double-stranded DNA is helical, and the two strands in the helix are
antiparallel. The double helix is stabilized by hydrogen bonds between
purine and pyrimidine bases on the opposite strands.
A on one strand pairs by two hydrogen bonds with T on the opposite strand,
or G pairs by three hydrogen bonds with C. The two strands of double-
helical DNA are, therefore, complementary.
Ribonucleic acid (RNA) most frequently occurs in single-stranded form.
The uracil base (U) replaces thymine base (T) in DNA, so the
complementary bases that determine the structure of RNA are A-U and C-
G.
The most general function of RNA is communication of DNA gene
sequences in the form of messenger RNA (mRNA) to ribosomes. These
processes are referred to as transcription and translation. mRNA (referred
to as +ssRNA) is transcribed as the RNA complement to the coding DNA
strand. This mRNA is then translated by ribosomes. The ribosomes, which
contain both ribosomal RNA (rRNA) and proteins, translate this message
Figure 2
3
into the primary structure of proteins via aminoacyl-transfer RNAs
(tRNAs).
The Eukaryotic Genome
The genome is the totality of genetic information in an organism. Nearly all
of the eukaryotic genome is carried on two or more linear chromosomes
separated from the cytoplasm within the membrane of the nucleus. Diploid
eukaryotic cells contain two homologues (divergent evolutionary copies) of
each chromosome.
Eukaryotic cells contain mitochondria and, in the case of plants,
chloroplasts. Within each of these organelles is a circular molecule of DNA
that contains a few genes whose function relates to that particular organelle.
Many yeast contain an additional genetic element, an independently
replicating 2-μm circle containing about 6.3 kbp of DNA. Such small
circles of DNA, termed plasmids, are frequently associated with
prokaryotes.

4
The Prokaryotic Genome
Most prokaryotic genes are carried on the bacterial chromosome. And with
few exceptions, bacterial genes are haploid.
The most prokaryotic genomes (>90%) consist of a single circular DNA
molecule. A few bacteria (eg, Brucella melitensis, Burkholderia
pseudomallei, and Vibrio cholerae) have genomes consisting of two
circular DNA molecules.
Many bacteria contain additional genes on plasmids that range in size from
several to 100 kbp. In contrast to eukaryotic genomes, 98% of bacterial
genomes are coding sequences.
Covalently closed DNA circles (bacterial chromosomes and plasmids),
which contain genetic information necessary for their own replication, are
called replicons or episomes.
Some bacterial species are efficient at causing disease in higher organisms
because they possess specific genes for pathogenic determinants. These
genes are often clustered together in the DNA and are referred to as
pathogenicity islands.
Transposons are genetic elements that contain several genes, including
those necessary for their migration from one genetic locus to another.
The short transposons (0.75–2.0 kbp long), known as insertion elements,
produces the majority of insertion mutations. These insertion elements
(also known as insertion sequence [IS] elements) carry only the genes for
enzymes needed to promote their own transposition to another genetic
locus but cannot replicate on their own.
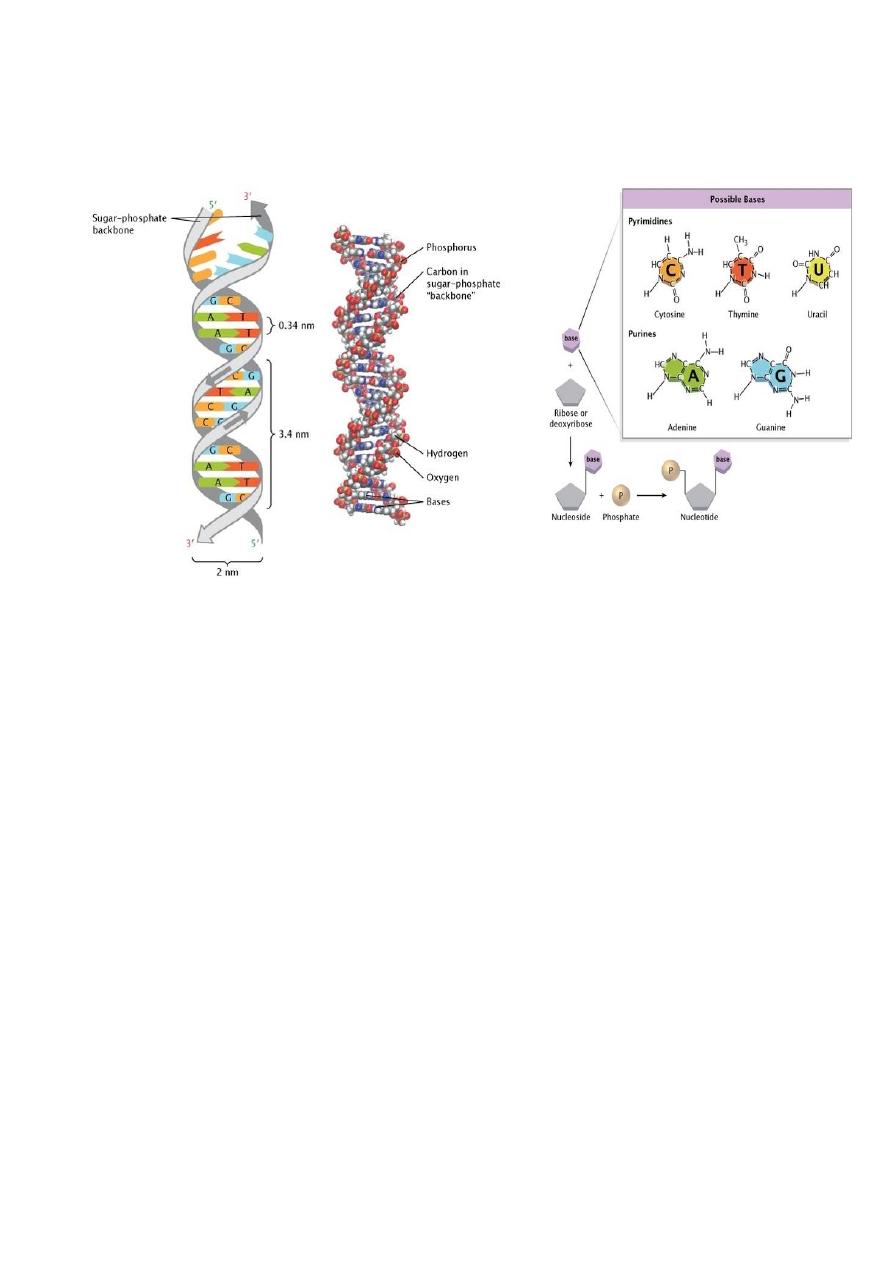
Replication
The replication of bacterial DNA begins at one point and moves in both
directions (bidirectional replication).
In the process, the two old strands of DNA are separated and used as
templates to synthesize new strands (semiconservative replication).
5
The structure where the two strands are separated and the new synthesis is
occurring is referred to as the replication fork.
Replication of the bacterial chromosome is controlled, and the number of
each chromosomes (when more than one is present) per growing cell falls
between one and four.
The replication of circular double-stranded bacterial DNA begins at the ori
locus and involves interactions with several proteins. In E coli, chromosome
replication terminates in a region called ter. The two daughter chromosomes
are separated, before cell division, so that each progeny cell gets one of the
daughter DNAs.
Similar processes used in the replication of bacterial chromosomes are used
in the replication of plasmid DNA.
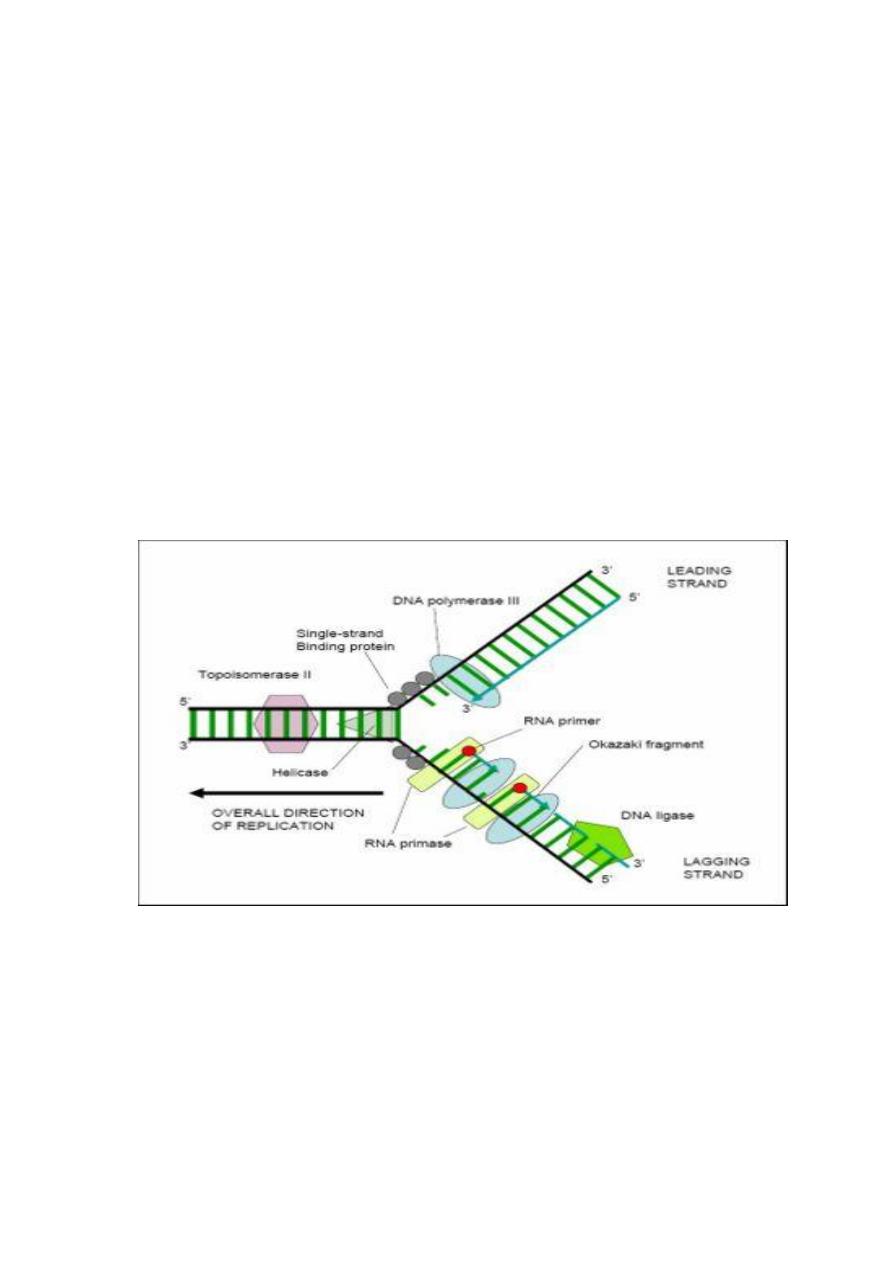
Gene expression
In both eukaryotic and prokaryotic genomes, genetic information is encoded
in DNA, transcribed into mRNA, and translated on ribosomes through
tRNA into the structure of proteins. The mechanism by which the sequence
of nucleotides in a gene determines the sequence of amino acids in a protein
is largely similar in prokaryotes and eukaryotes and is as follows:
6
1. RNA polymerase forms a single polyribonucleotide strand, called mRNA, using
DNA as a template; this process is called transcription. The mRNA has a
nucleotide sequence complementary to a template strand in the DNA double helix
if read in the 3′–5′ direction. Thus, an mRNA is oriented in a 5′–3′ direction.
2. Amino acids are enzymatically activated and transferred to specific adapter
molecules of RNA, called tRNA. Each adapter molecule has a triplet of bases
(anticodon) complementary to a triplet of bases on mRNA, and at one end its
specific amino acid. The triplet of bases on mRNA is called the codon specific for
that amino acid.
3. mRNA and tRNA come together on the surface of the ribosome. As each tRNA
finds its complementary nucleotide triplet on mRNA, the amino acid that it carries
is put into peptide linkage with the amino acid of the preceding tRNA molecule.
The ribosome moves along the mRNA with the nascent polypeptide growing
sequentially until the entire mRNA molecule has been translated into a
corresponding sequence of amino acids. This process, called translation.
Genes of higher eukaryotes are interrupted by introns, (long sequences that do not
code for proteins) which are spliced out of the RNA before translation (post-
transcription process).

7
Mechanisms of Gene Transfer
The maintains of genetic variability in microbes through the exchange and
recombination of allelic forms of genes. Recombination refers to the acquisition
of new segments of DNA from the local environment or provided by another
microorganism. When the recombination takes place, new genes or new
recombination are added to those already present in a microorganism.
DNA can be transferred from one organism to another, and that DNA can be
stably incorporated in the recipient, permanently changing its genetic composition,
this process is called horizontal gene transfer (HGT) to differentiate it from the
inheritance of parental genes, a process called vertical inheritance. Three broad
mechanisms mediate efficient movement of DNA between cells: conjugation,
transduction, and transformation.
Conjugation
is a one-way transfer of genetic material (usually plasmids) from donor to
recipient by means of physical contact. Conjugation requires donor cell to-
recipient cell contact to transfer only one strand of DNA.
The recipient completes the structure of dsDNA by synthesizing the strand
that complements the strand acquired from the donor. F
+
possesses a fertility
(F) plasmid, mediating the creation of a sex pilus necessary for conjugal
transfer of the F plasmid to the recipient.
It can integrate into chromosomal DNA, creating high-frequency
recombination (HFR) donors from which chromosomal DNA is readily
transferred. R factor contain genes conferring drug resistance. Frequently,
the resistance genes are carried on transposons. express resistance
phenotype through natural selection.
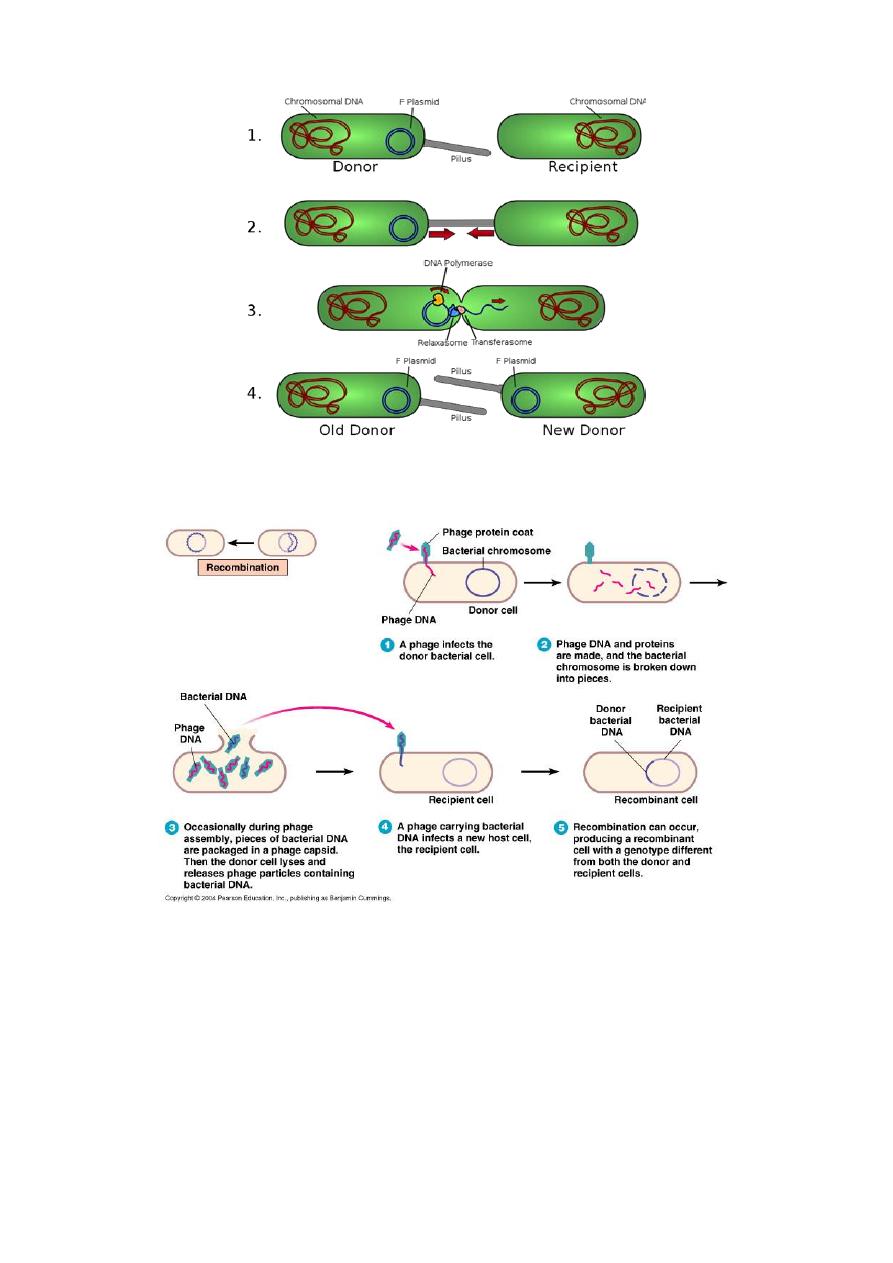
8
2- Transduction: is phage-mediated genetic recombination in bacteria. DNA is
carried by a phage coat and is transferred into the recipient cells.
3- Transformation: the direct uptake of “naked” donor DNA by the recipient
cell, may be natural or forced. Forced transformation is induced in the laboratory,
where, after treatment with high salt and temperature shock, many bacteria are
rendered competent for the uptake of extracellular plasmids.
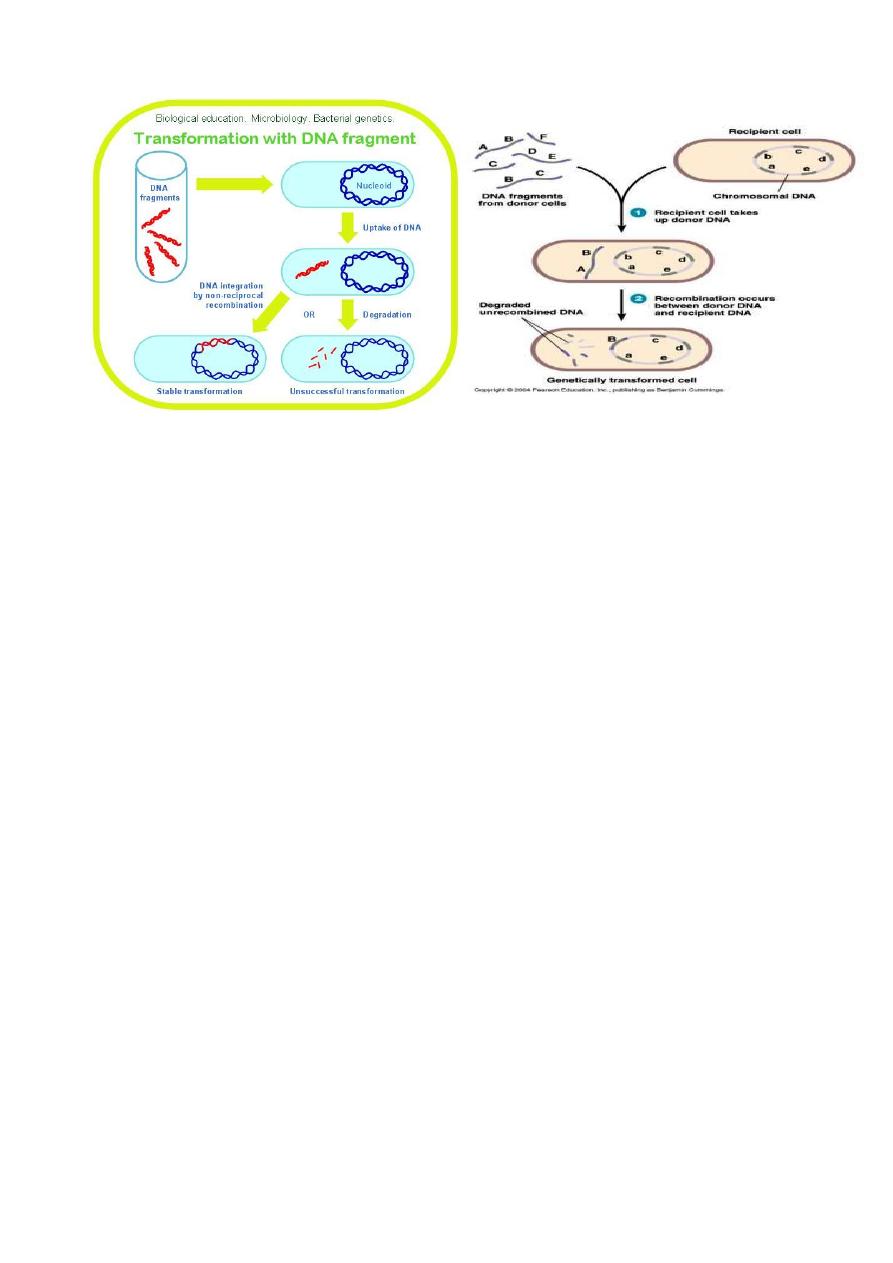
9
Mutation
is the permanent alteration of the nucleotide sequence of the genome of an
organism, virus, or extrachromosomal DNA or other genetic elements. Mutation
occurs approximately once for any gene in every 1 million cells.
It is an induced
or spontaneous heritable alteration of the DNA sequence. The spontaneous
mutation is a random change in the DNA arising from errors in replication that
occur without a known cause. Induced mutations result from exposure to known
mutagens, which are primarily physical or chemical agents that damage DNA and
interfere with its functioning. The frequency of spontaneous mutations has been
measured for a number of organisms. It introduces variability into the gene and
changes in the phenotype.
Mutation may be caused by various mutagens,
including ultraviolet light, acridine dyes, base analogues, and nitrous acid. The
mutations include base substitutions, deletions, insertions, and rearrangements.
Nucleotide substitutions: arise from mutagenic activity or the mis-pairing of
complementary bases during DNA replication. It's often do not significantly
disrupt the function of gene products.
Frameshift mutations: result from the insertion or deletion of one or two base
pairs, disrupting the phase of the triplet-encoded DNA message.
Rearrangements are the result of deletions that remove large portions of genes or
even sets of genes.
11
Deletions: are usually large excisions of DNA, dramatically altering the sequence
of coded proteins. It may also result in frameshift mutations.
Insertions: change genes and their products by integration of new DNA via
transposons.
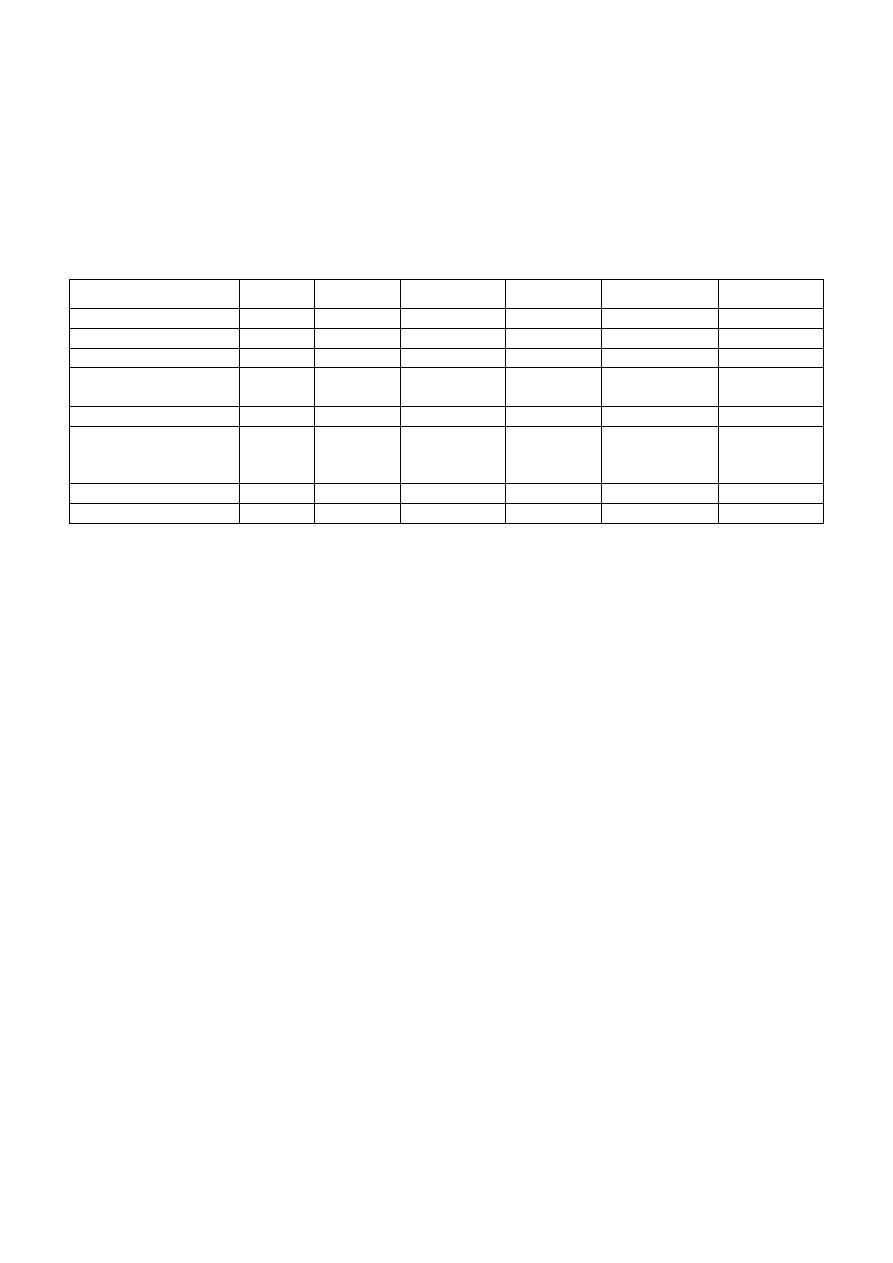
— The genetic systems (genome) of different living things
Structure
Virus
Bacteria
Fungus
Plant
Animal
Human
Nucleus
— -
— -
— +
— +
— +
— +
Nuclear membrane
— -
— -
— +
— +
— +
— +
chromosome
genome
single
multiple
multiple
multiple
multiple
Extra chromosome
— -
-
Plasmid
mitochondria chloroplast
mitochondria
mitochondria
mRNA
mature
mature
immature
immature
immature
immature
DNA
— +
/
-
— +
— +
— +
— +
— +
Intron
— -
— -
— +
— +
— +
— +
Histone
— -
— -
— +
— +
— +
— +