
1
Orthomyxoviruses
اﻟﻤﺮﺣﻠﮫ اﻟﺜﺎﻟﺜﮫ /ﻓﺎﯾﺮوﺳﺎت
د. اﻧﺘﻈﺎر ﻋﻼوي ﺟﻌﻔﺮ / ﻓﺮع اﻻﺣﯿﺎء اﻟﻤﺠﮭﺮﯾﮫ / ﻛﻠﯿﮫ اﻟﻄﺐ / ﺟﺎﻣﻌﮫ ذي ﻗﺎر
PhD. M.Sc. Microbiology
Introduction
All these viruses were grouped under myxovirus (myxa meaning mucus) due to
their affinity to mucins (glycoproteins on cell surface as their receptors).
These viruses are classified under two families
These are:
•
Orthomyxoviridae, consisting of influenza viruses.
•
Paramyxoviridae, consisting of parainfluenza, mumps, measles,
respiratory syncytial, and Newcastle disease viruses.
•
The genus Orthomyxovirus includes influenza viruses, the causative
agents of worldwide epidemics of influenza.
Influenza Viruses
Influenza viruses belong to the family of Orthomyxoviridae and are the causative
agents of influenza, a respiratory disease in humans with well-defined systemic
symptoms that occurs in sporadic, epidemic, and pandemic forms. Influenza A
and B viruses cause substantial morbidity and mortality in humans and a
considerable financial burden worldwide, whereas influenza C viruses cause
sporadic outbreaks of mild respiratory disease, mainly in children.
N.B
v
Epidemic refers to an increase, often sudden, in the number of cases of a disease
above what is normally expected in a population within a geographic area
v
Pandemic refers to an epidemic that has spread over several countries or
continents, usually affecting a large number of people.
Properties of the Virus
Morphology
Influenza viruses are spherical or filamentous, enveloped particles 80–120 nm in
diameter.
It is composed of a characteristic segmented single-stranded RNA genome, a
nucleocapsid, and an envelope (See-Fig).
Source: ViralZone:www.expasy.org/viralzone, SIB Swiss Institute of Bioinformatics)

2
The viral genome is a single-stranded antisense RNA. The genome consists of an
RNA-dependent RNA polymerase, which transcribes the negative-polarity
genome into mRNA.
• The RNA genome is segmented and consists of eight segments in
Influenza A & B and seven segments in influenza C viruses. These
segments code for different proteins, which are NS1, NS2, NP, M1, M2, M3,
HA, and NA.
The genome is present in a helically symmetric nucleocapsid surrounded by a
lipid envelope. The envelope has an inner membrane protein layer and an outer
lipid layer. The membrane proteins are known as matrix or M protein and are
composed of two components M1 and M2.
•
Two types of spikes or peplomers project from the envelope:
(a) The triangular hemagglutinin (HA) peplomers and
(b) The mushroom-shaped neuraminidase (NA) peplomers.
18 hemagglutinin (HA) and 11 NA subtypes of influenza A viruses are found in
nature.
Antigenic and genomic properties
•
Influenza viruses have two types of antigens:
•
Group-specific antigens: The ribonucleoprotein (RNP) antigen, or the
“soluble” antigen, is the group-specific antigen.
•
Influenza viruses are divided into types A, B, and C on the basis of
variation in this nucleoprotein antigen.
Type-specific antigens: The surface antigen, or “viral” antigen, or “V antigen” is
composed of two virus-encoded proteins, Hemagglutinin (HA) and NA, which are
the type-specific antigens.
Hemagglutinin (HA)
HA is a trimer (is a molecule formed by association of three molecules of the
same substance) and is composed of two polypeptides, HA1 and HA2,
responsible for hemadsorption and hemagglutination. The hemagglutinin
consists of 500 spikes, the triangular-shaped HA is inserted into the virus
membrane by its tail end. The distal end, which contains five antigenic sites
(designated as HA1–HA5), is responsible for binding of virion to host cells.
Influenza viruses adsorb many avian and mammalian erythrocytes.
Hemagglutinin binds with the neuraminic acid (sialic acid) cell receptor, and
initiates the infection in the host cell. Adsorption of erythrocytes occurs at 4°C,
but at 37°C there is detachment of the red cells due to destruction of the
glycoprotein receptors by the viral enzyme, neuraminidase.
The hemagglutinin agglutinates certain red blood cells, which is inhibited by the
neutralizing antibodies. This forms the basis of the hemagglutination inhibition
test used in the serodiagnosis of influenza.
Hemagglutinin has potency to undergo antigenic variations.
The nucleotide and amino acid sequences of the polypeptides, HA1 and HA2,
undergo radical changes in antigenic shift.

3
In antigenic drift, only minor changes take place in the compositions of HA
antigenic sites.
Neuraminidase (NA)
The NA is a glycoprotein and tetramer. It consists of 100 mushroom-shaped
spikes. The NA is inserted into the virus membrane by its hydrophobic tail end.
The distal end contains antigenic as well as enzymatically active sites. The NA
causes hydrolysis of red cells, hence causes elution or detachment of the cells
adsorbed to virion particles. The function of the neuraminidase is to cleave the
neuraminic acid and to release progeny virions from the infected host cells.
Antigenic variations
Antigenic variation is a unique feature of influenza virus.
The surface antigens HA and NA show variations and are primarily responsible
for antigenic variations exhibited by influenza viruses. The internal RNP antigen
and M protein are stable, hence do not contribute to the antigenic variations.
Antigenic variations are of two types: antigenic shift and antigenic drift.
Antigenic shift
Antigenic shift: The abrupt, drastic, discontinuous change. This occurs due to
major antigenic changes in HA or NA antigens, and is caused by replacement of
the gene for HA by one coding for a completely different amino acid sequence.
The antigenic shift is characterized by alteration of virtually all the antigenic
sites of the HA.
Antigenic drift
Is gradual, sequential, regular antigenic change in influenza virus. This occurs
due to minor antigenic changes in the HA or NA occurring at frequent intervals.
This is caused even by a single mutation affecting HA glycoprotein. The antigenic
drift is characterized by changes in certain epitopes in the HA, while others are
being conserved.
Table: Differences between antigenic shift and antigenic drift
4
Key Points
•
Influenza A virus shows maximum antigenic variations.
•
Influenza B virus does not undergo antigenic shift because influenza B
virus is the only human virus for which there is no animal source of new
RNA segments. However, influenza B virus undergoes antigenic drift.
•
Antigenic variation never occurs in type C influenza virus beacuse its lack
of NA.
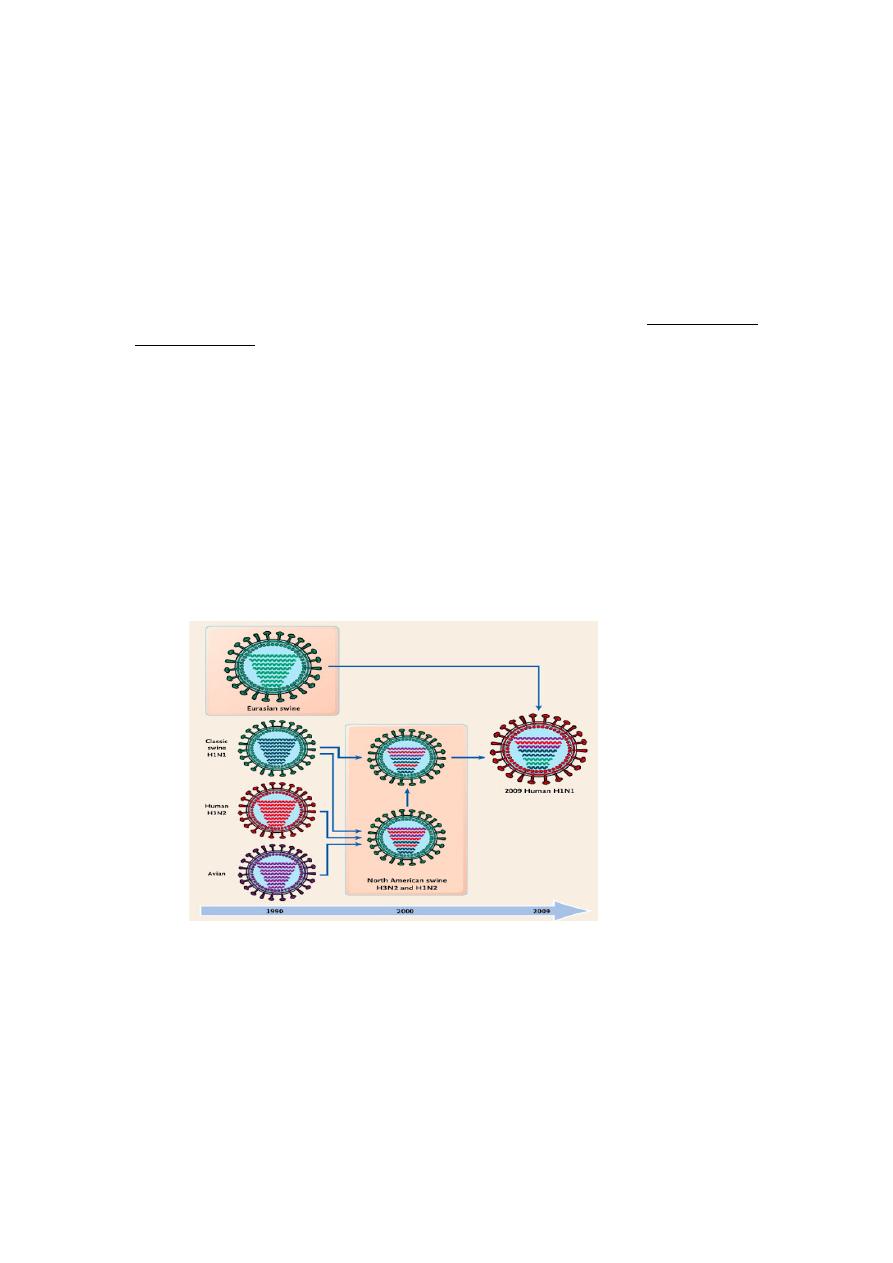
Gene reassortment
Because the influenza virus genome is segmented, genetic reassortment can
occur when a host cell is infected simultaneously with viruses of two different
parent strains. This process of genetic reassortment accounts for the periodic
appearance of the novel types of influenza A strains that cause influenza
pandemics.
Influenza viruses of animals, such as aquatic birds, chickens, swine, and horses
show high host specificity. These animal viruses are the source of the RNA
segments that encode the antigenic shift variants that cause epidemics among
humans. For example, if a person is infected simultaneously by an avian and
human influenza strains, then it is possible that a genetic reassortment could
occur in infected cells in humans. The reassortment could lead to emergence of a
new human influenza A virus, the progeny of which will contain a mixture of
genome segments from the two strains (e.g., a new variant of human influenza A
virus bearing the avian virus HA).
Designation of influenza viruses
Influenza virus type A can be classified into subtypes based on the variations in
their surface antigens. The WHO proposed a new system of classification in 1971
and was later modified, which takes into account the nature of both the surface
antigens. According to this, the complete designation of a strain will include the
(a) type, (b) place of origin, (c) serial number, and (d) year of isolation followed
by (e ) antigenic subtypes of the HA and NA in parentheses.
5
For example: influenza A/Singapore/1/57 (H2N2) indicates that influenza was
first originated from Singapore and was isolated for the first time in the year
1957. The HA and NA antigens are H2 and N2 as shown in the parentheses.
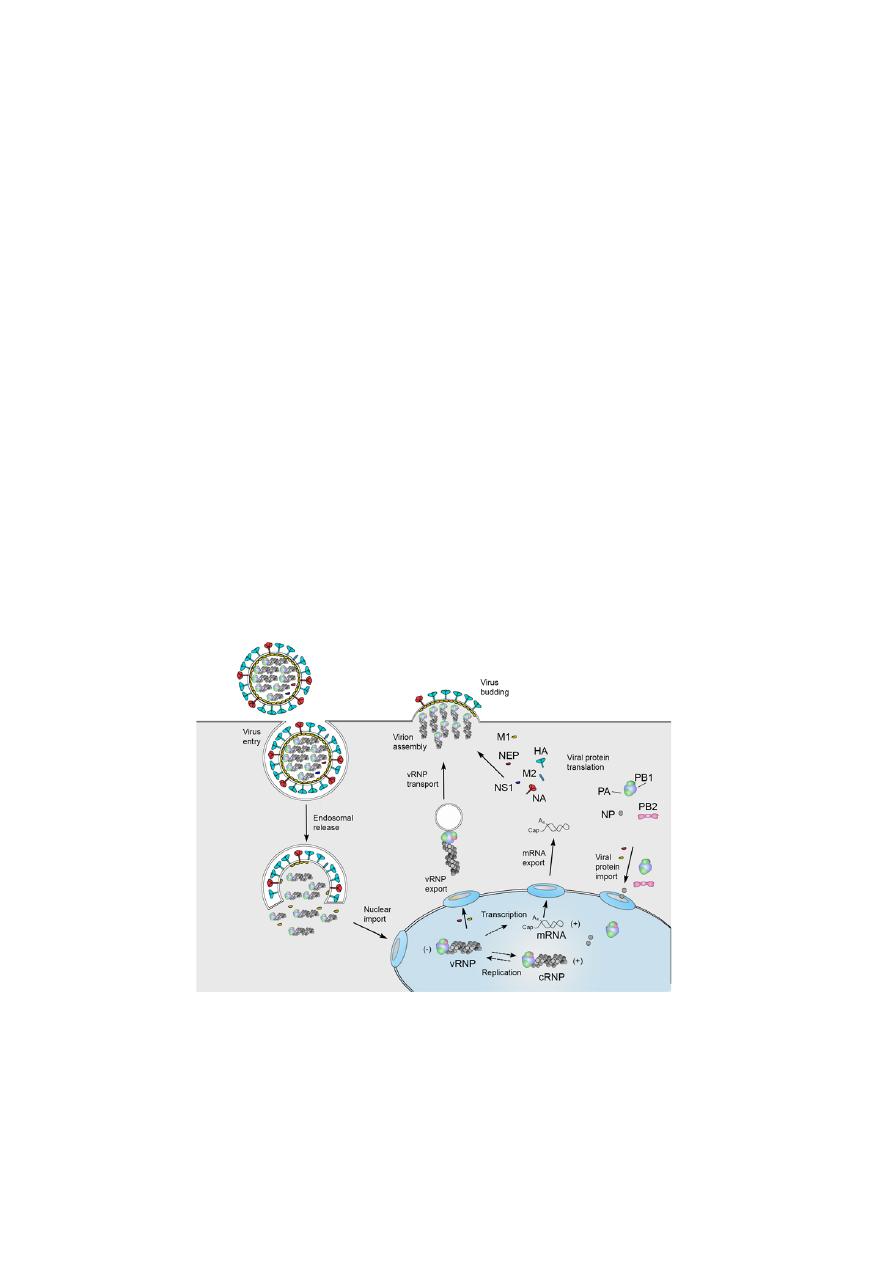
REPLICATION
•
Viral infection initiates with the binding of a virion to cell surface
receptors containing sialic acid, followed by the endocytosis of the virion.
•
After fusion of the viral and endosomal membranes, the viral
ribonucleoproteins (vRNPs) are released into the cytoplasm and then
transported into the nucleus.
•
In the nucleus the viral RNA polymerase transcribes the vRNA segments
into mRNAs.
•
Viral mRNA is exported to the cytoplasm for translation by cellular
mechanisms. The viral RNA polymerase also performs replication of
vRNA by copying it into complementary RNA (cRNA), which serves as a
template for the production of more vRNA. Newly synthesised viral
polymerase and nucleoprotein are imported into the nucleus and bind to
cRNA and vRNA to assemble vRNPs and cRNPs, respectively.
•
Following nuclear export, progeny vRNPs are transported across the
cytoplasm on recycling to the cell membrane, where assembly of progeny
virions takes place.
•
Mature virions are released by budding.
Infleunza A virus replication cycle.
Source:
Aartjan J.W. te Velthuis and Ervin Fodor, 2017.

6
Pathogenesis and Immunity
Influenza virus is transmitted from person to person primarily in droplets
released by sneezing and coughing.
Inhaled influenza viruses reach lower respiratory tract, tracheobronchial tree,
the primary site of the disease. They attach to sialic acid receptors on epithelial
cells by HA present on the viral envelope. Relatively few viruses are needed to
infect lower respiratory tract than the upper respiratory tract. Neuraminidase of
the viral envelope may act on the N -acetyl neuraminic acid residues in mucus to
produce liquefaction.
Infection of mucosal cells results in cellular destruction and desquamation of the
superficial mucosa. The resulting edema and mononuclear cell infiltration of the
involved areas are accompanied by symptoms including nonproductive cough,
sore throat, and nasal discharge. Although the cough may be striking, the most
prominent symptoms of influenza are systemic: fever, muscle aches, and general
prostration. The virus remains localized to the respiratory tract; hence viremia
does not occur. In an uncomplicated case, virus can be recovered from
respiratory secretions for 3–8 days.
Clinical Syndrome
Incubation period is short (1–3 days). The classic influenza syndrome is a febrile
illness of sudden onset, characterized by tracheitis and marked myalgias.
Headache, chills, fever, malaise, myalgias, anorexia, and sore throat appear
suddenly. The body temperature rapidly rises to (38.3–40.0°C) and respiratory
symptoms ensue. Nonproductive cough is characteristic. Sneezing, rhinorrhea,
and nasal obstruction are common.
Patients may also report photophobia, nausea, vomiting, diarrhea, and
abdominal pain. They appear acutely ill and are usually coughing. Minimal to
moderate nasal obstruction, nasal discharge, and pharyngeal inflammation may
be present.
Complications
1-Secondary bacterial infections: Life-threatening influenza is often caused by
secondary bacterial infections with staphylococci, pneumococci, and
Haemophilus influenzae. Pneumonia may develop as a complication and may be
fatal, particularly in
(a) Elderly persons above 60 years with underlying chronic disease.
(b) In people chronic cardiorespiratory disease, renal disease, etc.)
(c) Pregnant women.
2-Reye’s syndrome is a noted complication of influenza B infection. The
condition is seen most commonly in young children and is associated with
degenerative changes in the brain, liver, and kidney.
3- Central nervous system complications: Guillain–Barre syndrome
characterized by encephalomyelitis and polyneuritis is a rare complication of
influenza virus infection.

7
Influenza epidemics and pandemics
Influenza epidemics are of two types. Yearly epidemics are caused by both type A
and type B viruses. The rare, severe influenza pandemics are always caused by
type A virus. Antigenic shift and antigenic drift are the two different mechanisms
responsible for producing the strains that cause these two types of epidemics.
Antigenic shift
A major change in one or both of the surface antigens, a change that yields an
antigen showing no serologic relationship with the antigen of the strains
prevailing at the time is called antigenic shift. Antigenic shift has been
demonstrated in type A influenza virus only.
Antigenic shift variants appear less frequently, about every 10 or 11 years. It is
demonstrated that pandemic strains are the recombinant strains, originated
from some animal or bird reservoir, either spreading to humans directly by
host range mutation or as a result of a recombination between human and
nonhuman strains. The completely novel antigens that appear during antigenic
shift are acquired by genetic reassortment.
The donor of the new antigens is probably an animal influenza virus. Type A
viruses have been identified in pigs, horses, and birds, and animal influenza
viruses possessing antigens closely related to those of human viruses. Fourteen
distinct HA and nine NA antigens are known.
Antigenic drift
Repeated minor antigenic changes, on the other hand, generate strains that
retain a degree of serologic relationship with the currently prevailing strain. The
epidemics are caused by influenza A virus undergoing antigenic variations due to
antigenic drift resulting from mutations and selections. Antigenic drift variants
occur very frequently, virtually every year. This is responsible for emergence of
the strains that cause yearly influenza epidemics.
Reservoir, source, and transmission of infection
Infected humans are the main reservoir of infections for influenza A virus.
Respiratory secretions of infected persons are the important source of infection.
The virus is excreted in respiratory secretions immediately before the onset of
illness and for 3–4 days thereafter. Wild aquatic birds are known reservoirs of
influenza A. They secrete the viruses in their feces, which contaminates ponds
and lakes. The virus is spread from person-to-person primarily by air-borne
respiratory droplets released during the acts of sneezing and coughing.
Influenza B virus only causes epidemics. Infection is from humans-to-humans.
No animal reservoir hosts are known.
Laboratory Diagnosis
During an epidemic of influenza, the clinical diagnosis can be made, but
definitive diagnosis depends on the laboratory methods.
Specimens:
•
Nasal or throat washings or sputum for viral antigen and viral RNA.
•
Throat gargles for isolation of viruses.
8
•
Serum for viral antibodies.
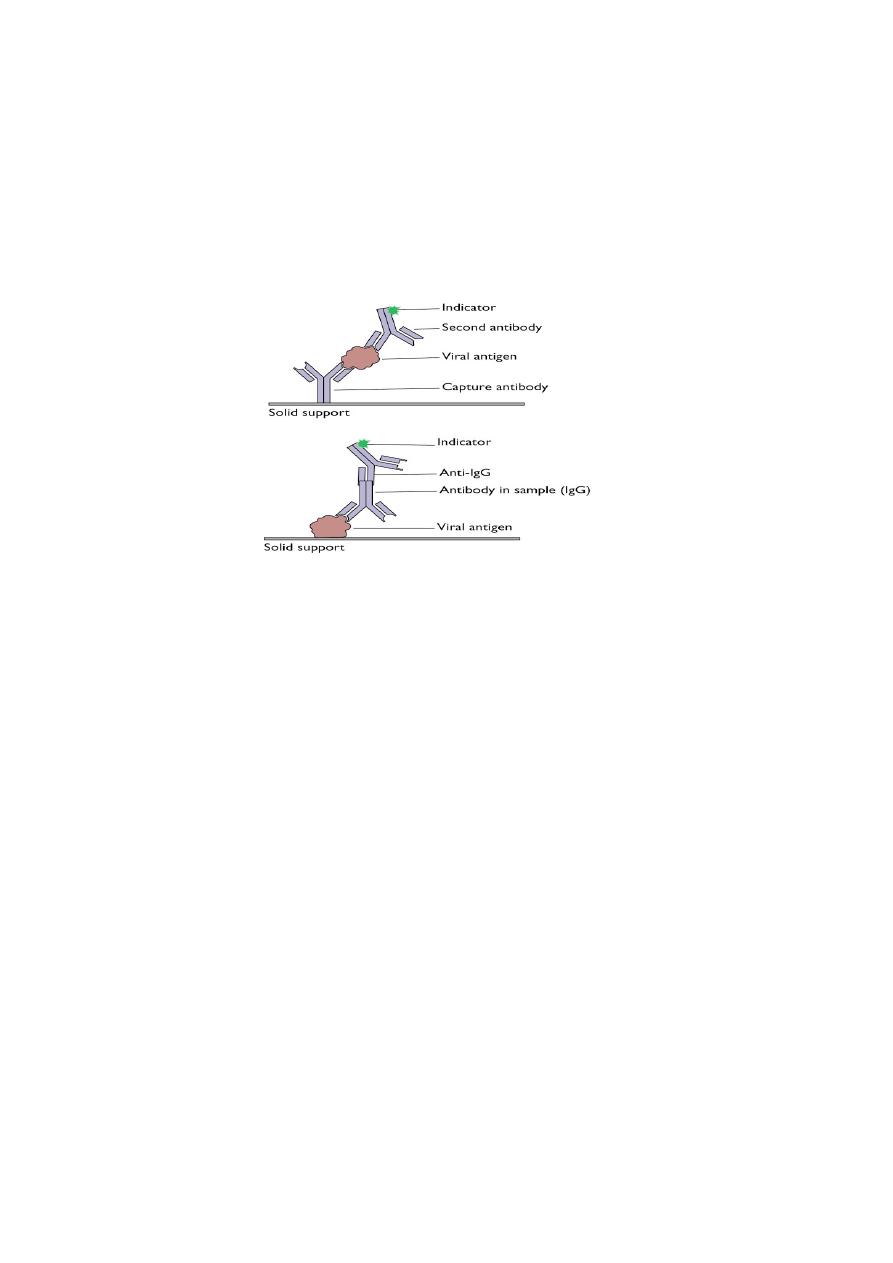
Direct antigen detection
Is made by demonstrating viral antigens directly on cells obtained from the
nasopharynx. Immunofluorescence (IF) or enzyme-linked immunosorbent assay
using specific monoclonal antibodies are used to detect viral antigen.
The results of the rapid tests are useful to start treatment with the NA inhibitors
within 48 hours of the onset of symptoms.
Detection of antigens by ELISA
Isolation of the virus
Throat gargles are the specimen of choice. The specimen is collected in saline
broth or a buffered salt solution and is sent immediately to the laboratory, or if
delayed is stored at 4°C.
The virus is isolated from the specimen by inoculation into embryonated eggs or
into certain cell cultures.
Treatment
Amantadine and Rimantadine are the specific antiviral agents available for
treatment of influenza.
Prevention and Control
This is based on the following:
Immunoprophylaxis by vaccines : Influenza A subtypes H1N1 and H3N2 are
most common predominate human influenza viruses. The trivalent vaccine used
worldwide contains influenza A strains from H1N1 and H3N2, along with an
influenza B strain.
