
1
3
rd
Lecture Medical students Medical Biology
Genetic Material
In this section, we will focus on some techniques used in molecular genetics.
Nucleic acid hybridization:
Extremely sensitive detection method, capable of picking out specific DNA
sequences from complex mixtures. Usually a single pure sequence is labeled with P³² and
used as a probe. The probe is denatured before use so that the strands are free to base-pair
with their complements. When DNA is hybridized with a RNA probe, the DNA-RNA
hybrids are immobilized onto a plate using anti-DNA-RNA antibodies. Antihybrid
antibodies conjugated to an enzyme are used for detection in combination with appropriate
chemiluminescent substrates.
Gel electrophoresis:
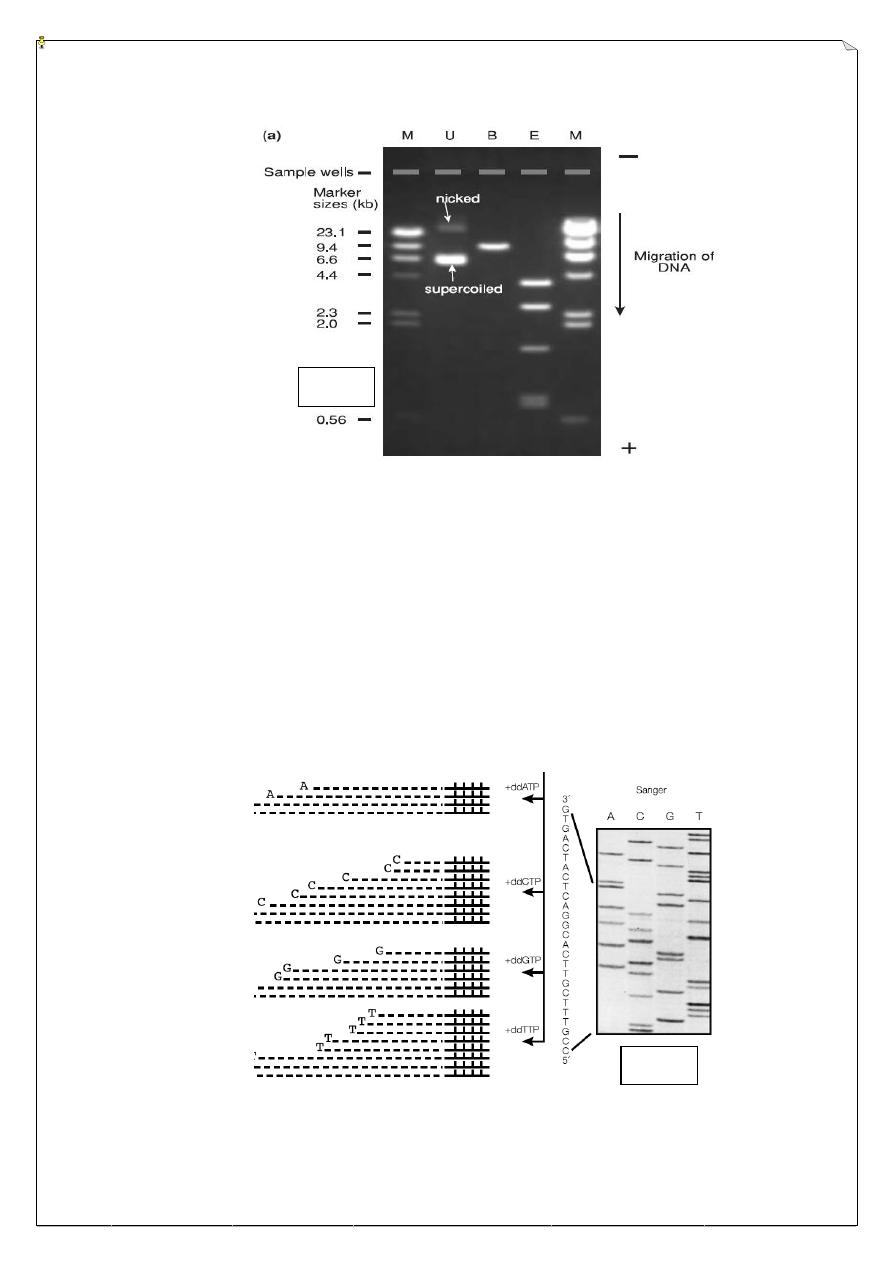
The technique of gel electrophoresis is vital to the genetic engineer, as it
represents the main way by which nucleic acid fragments may be visualized directly.
When an electric field is applied to an agarose gel in the presence of a buffer solution
which will conduct electricity, DNA fragments move through the gel towards the positive
electrode (DNA is highly negatively charged) at a rate which is dependent on their size
and shape. Hence, this process of electrophoresis may be used to separate mixtures of
DNA fragments on the basis of size. The DNA samples are placed in wells in the gel
surface. DNA is allowed to migrate through the gel in separate lanes. The DNA is stained
by the inclusion of ethidium bromide. After electrophoresis, The DNA shows up as an
orange band on illumination by UV light.
2
DNA sequencing:
The ability to determine the sequence of bases in DNA is a central part of modern
molecular biology and provides what might be considered the ultimate structural
information. The two main methods of DNA sequencing are the Maxam and Gilbert
chemical method in which end-labeled DNA is subjected to base-specific cleavage
reactions prior to gel separation, and Sanger’s enzymic method. If, however, the aim is to
sequence a much larger piece of DNA (such as an entire chromosome), the problem is
much greater.
لالطالع
لالطالع

3
The polymerase chain reaction:
The polymerase chain reaction (PCR) is used to amplify a sequence of DNA using
a pair of oligonucleotide primers each complementary to one end of the DNA target
sequence. These are extended towards each other by a thermostable DNA polymerase in a
reaction cycle of three steps: denaturation, primer annealing and polymerization. The
reaction cycle comprises a 95°C step to denature the duplex DNA, an annealing step of
around 55°C to allow the primers to bind and a 72°C polymerization step. Mg 2+ and
dNTPs are required in addition to template primers, buffer and enzyme.
Enzyme-linked immunosorbent assay (ELISA):
ELISA is a test that uses antibodies and color change to identify a substance
(antigen or antibody).Antigens from the sample are attached to a surface. Then, a further
specific antibody is applied over the surface so it can bind to the antigen. This antibody is
linked to an enzyme, and, in the final step, a substance containing the enzyme's substrate
is added. The subsequent reaction produces a detectable signal, most commonly a color
change in the substrate.
Southern and northern blotting:
The nucleic acid in lanes of a gel is transferred to a membrane, bound and then
hybridized with a labeled nucleic acid probe. Washing removes non hybridized probe, and
the membrane is then treated to reveal the bands produced. Specific RNA species are
detected on Northern blots, whereas the DNA bands on Southern blots could be genes in
genomic DNA or parts of cloned genes.
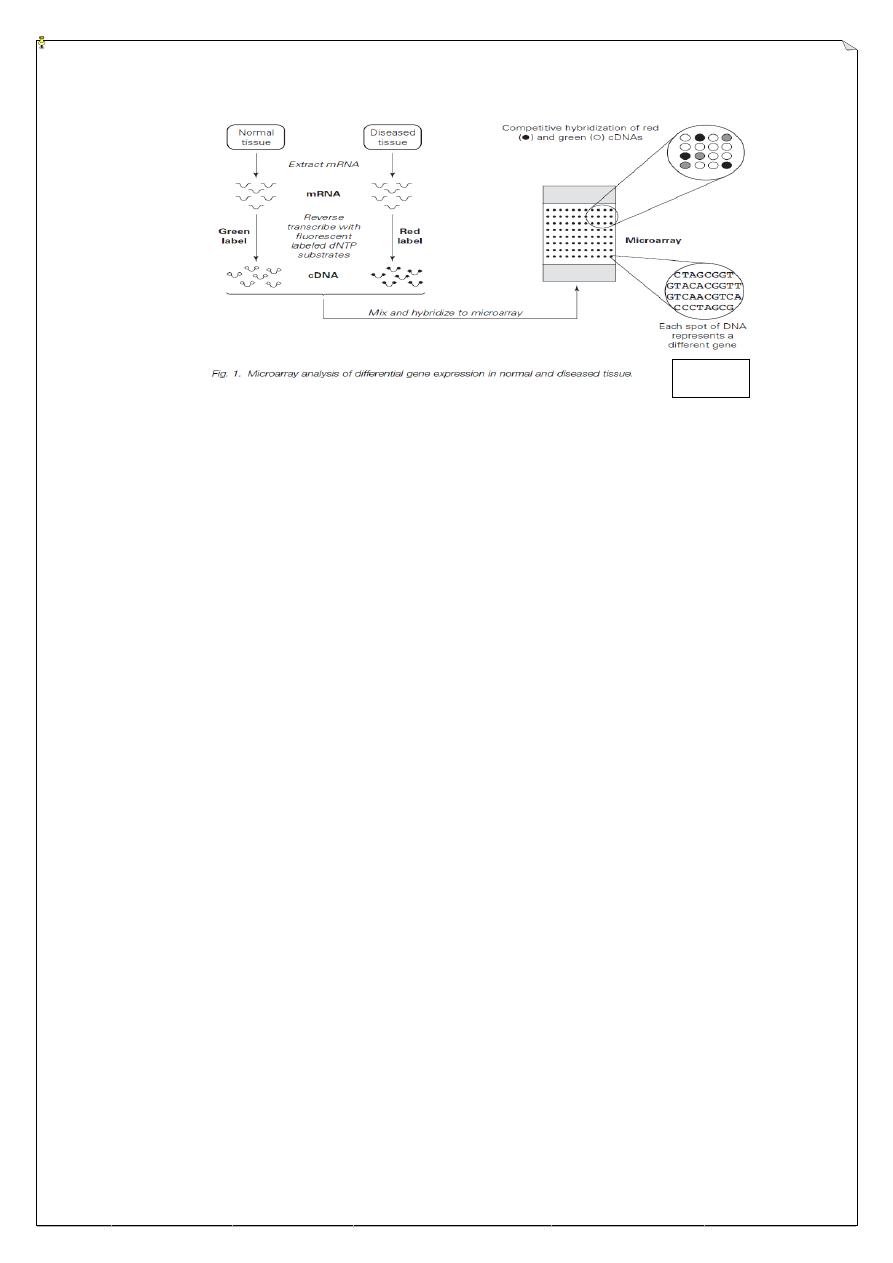
DNA Microarray:
DNA microarrays are small, solid supports (e.g. glass slides) on to which are
spotted individual DNA samples corresponding to every gene in an organism. When
hybridized to labeled cDNA (complementary) representing the total mRNA population of
a cell, each cDNA (mRNA) binds to its corresponding gene DNA and so can be separately
quantified. Commonly, a mixture of cDNAs labeled with two different fluorescent tags
and representing a control and experimental condition are hybridized to a single array to
provide a detailed profile of differential gene expression.
4
DNA fingerprinting:
Cloning and genomic sequencing projects have identified many repetitive
sequences in the human genome. Some of these are simple nucleotide repeats that vary in
number between individuals but are inherited (VNTRs). If Southern blots of restriction
enzyme-digested genomic DNA from members of a family are hybridized with a probe
that detects one of these types of repeats, each sample will show a set of bands of varying
lengths. Some of these bands will be in common with those of the mother and some with
those of the father and the pattern of bands will be different for an unrelated individual.
This is the technique of DNA fingerprinting which is used in forensic science to
eliminate the innocent and convict criminals. It is also applied for maternity and paternity
testing in humans.
Gene therapy
:
Attempts have been made to treat some genetic disorders by delivering a normal
copy of the defective gene to patients. This is known as gene therapy. For some disorders,
the bone marrow is destroyed and replaced with treated cells that have had a normal gene,
or a protective gene. Gene therapy will produce transgenic somatic cells in the patient.
Gene therapy is in its infancy, but it seems to have great potential.
Bioinformatics:
Bioinformatics is the interface between biology and computer science. It comprises
the organization of many kinds of large-scale biological data, particularly that based on
لالطالع
5
DNA and protein sequence, into databases (normally Web-accessible), and the methods
required to analyze these data.
REGULATION OF GENE TRANSCRIPTION
Promoters
The promoter region of a gene is usually several hundred nucleotides long and
immediately upstream from the transcription initiation site. The promoter constitutes the
binding site for the enzyme machinery that is responsible for the transcription of DNA to
RNA, the RNA polymerase. Promoters often contain the consensus sequence 5´-TATA-3´
box, 30 to 50 base pairs upstream of the site at which transcription begins in prokaryote.
Many eukaryotic promoters also have a so-called CAAT box with a GGNCAATCT
consensus sequence centered about 75 base pairs upstream of the initiation start site. In
general, the promoter region has a high importance for the regulation of expression of
any gene.
Enhancers:
These are sequence elements which can activate transcription from thousands of
base pairs upstream or downstream. They may be tissue-specific or ubiquitous in their
activity and contain a variety of sequence motifs. There is a continuous spectrum of
regulatory sequence elements which span from the extreme long-range enhancer elements
to the short-range promoter elements.
لالطالع

6
Operators
Operators are nucleotide sequences that are positioned between the promoter and
the structural gene. They constitute the region of DNA to which repressor proteins bind
and thereby prevent transcription. Repression of transcription is accomplished by the
repressor protein attaching to the operator sequence downstream of the promoter sequence
(the point of attachment of the RNA polymerase). The enzyme must pass the operator
sequence to reach the structural genes start site. The repressor protein bound to the
operator physically prevents this passage and, as a result, transcription by the polymerase
cannot occur. Repressor proteins themselves can be affected by a variety of other proteins
or small molecules.
Biotechnology
is defined as “any technological application that uses biological systems, living
organisms, or derivatives thereof, to make or modify products or processes for specific
use”. Several branches of industry rely on biotechnological tools for the production of
food, beverages, pharmaceuticals and biomedicals. The techniques used for this purpose
include:
» isolation of the gene coding for a protein of interest.
» cloning (i.e. transfer) of this gene into an appropriate production host.
» improving gene and protein expression by using stronger promoters, improving
fermentation conditions etc. Together, these techniques are known as recombinant DNA
technology.
Up By Fahad A.
