محاضرات الكيمياء الحياتية كلية طب الاسنان/ جامعة كركوك المرحلة الثانية د. نوال عبدالله مرتضى Enzymes
Chemical Nature of Enzymes
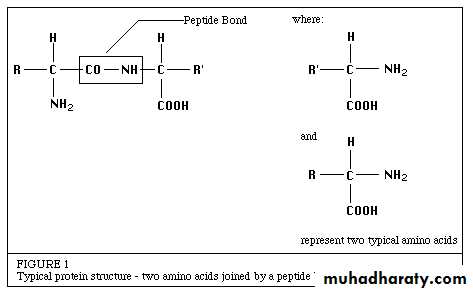
All known enzymes are proteins. They are high molecular weight compounds (biologic polymers) made up principally of chains of amino acids linked together by peptide bonds. See Figure 1.
Enzymes catalyze the chemical reactions that make life as we know it possible. It can be denatured and precipitated with salts, solvents and other reagents. They have molecular weights ranging from 10,000 to 2,000,000.
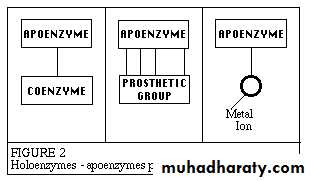
Many enzymes require the presence of other compounds - cofactors - before their catalytic activity can be exerted. This entire active complex is referred to as the holoenzyme; i.e., apoenzyme (protein portion) plus the cofactor (coenzyme, prosthetic group or metal-ion-activator) is called the holoenzyme.
Apoenzyme + Cofactor = Holoenzyme
Apo enzymeApo protein that forms an active enzyme system by combination with a coenzyme and determines the specificity of this system for a substrate
The cofactor may be:
1- A coenzyme - a non-protein organic substance which is dialyzable, thermo stable and loosely attached to the protein part.
2- A prosthetic group - an organic substance which is dialyzable and thermo stable which is firmly attached to the protein or apoenzyme portion.
3- A metal-ion-activator - these include K+, Fe++, Fe+++, Cu++, Co++, .Zn++, Mn++, Mg++, Ca++ and Mo +++.
Mode of action :
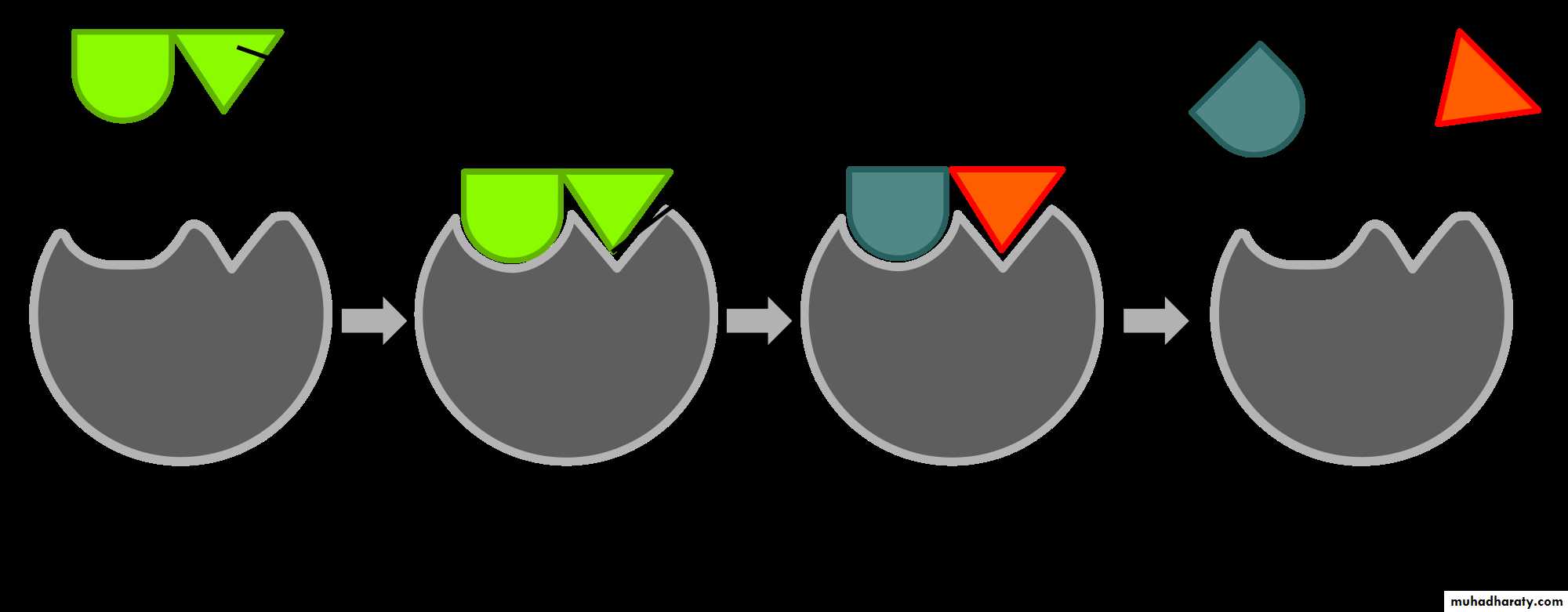
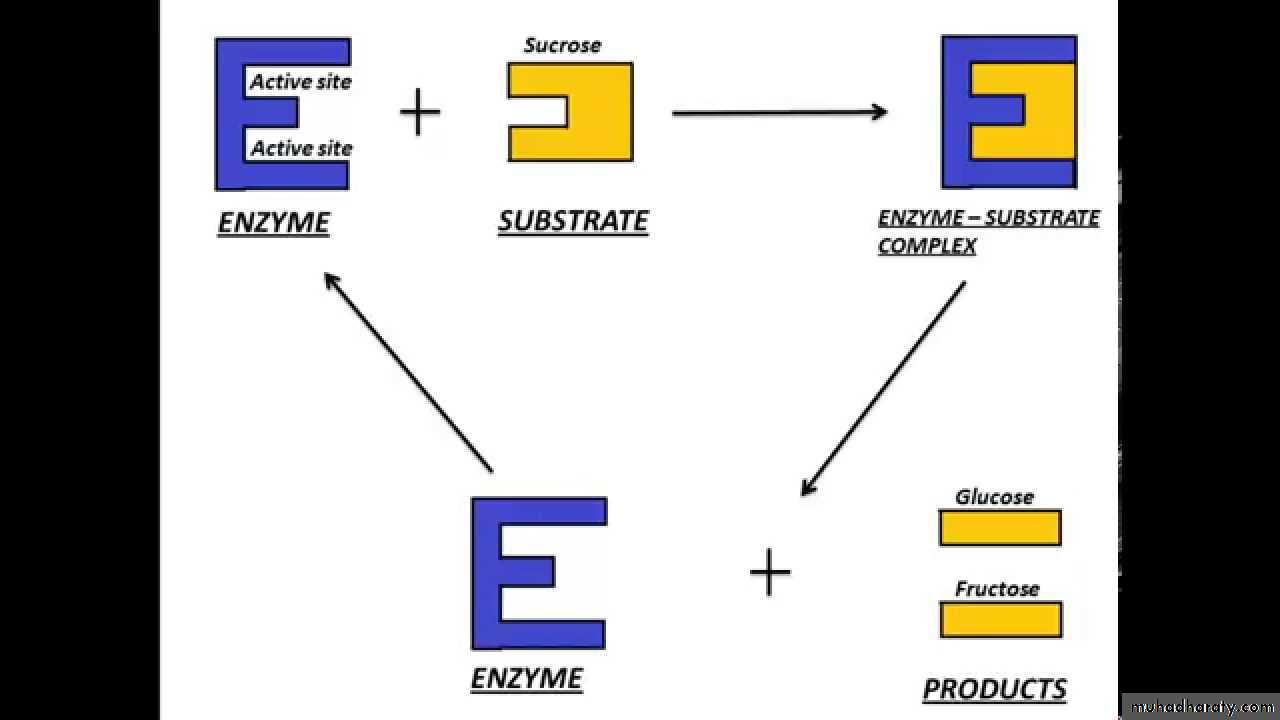
Enzymes enable chemical reactins to occur at cooler temperatures by reducing the amount of activation energy required to break the bonds of the reactant molecules. Each enzyme is very selective in the reaction it catalizes, this feature is based on the ability of the enzyme to recognize the shape of the certain reactant molecule - substrate. There is a special region on the enzyme that has same shape and chemistry as the substrate, so the substrate fits perfectly into tis docking station, the enzyme embraces it slightly and catalyzes the reaction. When the product is released, the enzyme is ready to accept another molecule of particular shape.
There are two common models of enzyme action. In the first model, the lock-and-key model, a protein called an enzyme "the lock" binds with another substance called a substrate "the key" and causes the lock to break down after forming an enzyme-substrate complex. The model is so named because substrates are very specific to individual enzymes. The second model is called the induced-fit model. It is pretty much the same as the first, except it is understood that the enzyme undergoes a conformational change before binding with the substrate. Enzymes are able to facilitate reactions at lower than usual temperatures because they lower the activation energy requirements of many biological reactions, acting as organic catalysts. This catalysis often requires "helper" substances called co-enzymes and cofactors. The rate at which enzymes work is determined by the type of enzyme, the amount of substrate present, and the amount of enzyme present. Some molecules can inhibit enzyme function by imitating the substrate or causing the enzyme to change shape.
Mechanism
Substrate binding
Enzymes must bind their substrates before they can catalyse any chemical reaction. Enzymes are usually very specific as to what substrates they bind and then the chemical reaction catalysed. Specificity is achieved by binding pockets with complementary shape, charge and hydrophilic/hydrophobic characteristics to the substrates. Enzymes can therefore distinguish between very similar substrate molecules to be chemoselective, regioselective and stereospecific.[26]
Some of the enzymes showing the highest specificity and accuracy are involved in the copying and expression of the genome. Some of these enzymes have "proof-reading" mechanisms. Here, an enzyme such as DNA polymerase catalyzes a reaction in a first step and then checks that the product is correct in a second step.[27] This two-step process results in average error rates of less than 1 error in 100 million reactions in high-fidelity mammalian polymerases.[1]:5.3.1 Similar proofreading mechanisms are also found in RNA polymerase,[28] aminoacyl tRNA synthetases[29] and ribosomes.[30]
Conversely, some enzymes display enzyme promiscuity, having broad specificity and acting on a range of different physiologically relevant substrates. Many enzymes possess small side activities which arose fortuitously (i.e. neutrally), which may be the starting point for the evolutionary selection of a new function.[31][32]
Enzyme changes shape by induced fit upon substrate binding to form enzyme-substrate complex. Hexokinase has a large induced fit motion that closes over the substrates adenosine triphosphate and xylose. Binding sites in blue, substrates in black and Mg2+ cofactor in yellow. (PDB: 2E2N, 2E2Q)
"Lock and key" model
To explain the observed specificity of enzymes, in 1894 Emil Fischer proposed that both the enzyme and the substrate possess specific complementary geometric shapes that fit exactly into one another.[33] This is often referred to as "the lock and key" model.[1]:8.3.2 This early model explains enzyme specificity, but fails to explain the stabilization of the transition state that enzymes achieve.[34]
Induced fit model
In 1958, Daniel Koshland suggested a modification to the lock and key model: since enzymes are rather flexible structures, the active site is continuously reshaped by interactions with the substrate as the substrate interacts with the enzyme.[35] As a result, the substrate does not simply bind to a rigid active site; the amino acid side-chains that make up the active site are molded into the precise positions that enable the enzyme to perform its catalytic function. In some cases, such as glycosidases, the substrate molecule also changes shape slightly as it enters the active site.[36] The active site continues to change until the substrate is completely bound, at which point the final shape and charge distribution is determined.[37] Induced fit may enhance the fidelity of molecular recognition in the presence of competition and noise via the conformational proofreading mechanism.[38Specificity of Enzymes
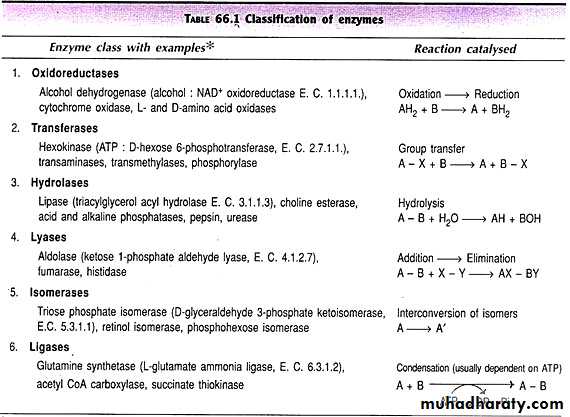
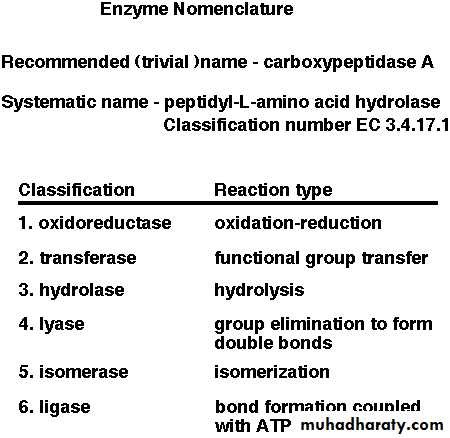
One of the properties of enzymes that makes them so important as diagnostic and research tools is the specificity they exhibit relative to the reactions they catalyze. A few enzymes exhibit absolute specificity; that is, they will catalyze only one particular reaction. Other enzymes will be specific for a particular type of chemical bond or functional group. In general, there are four distinct types of specificity:
Absolute specificity - the enzyme will catalyze only one reaction.
Group specificity - the enzyme will act only on molecules that have specific functional groups, such as amino, phosphate and methyl groups.Linkage specificity - the enzyme will act on a particular type of chemical bond regardless of the rest of the molecular structure.
Stereochemical specificity - the enzyme will act on a particular steric or optical isomer.
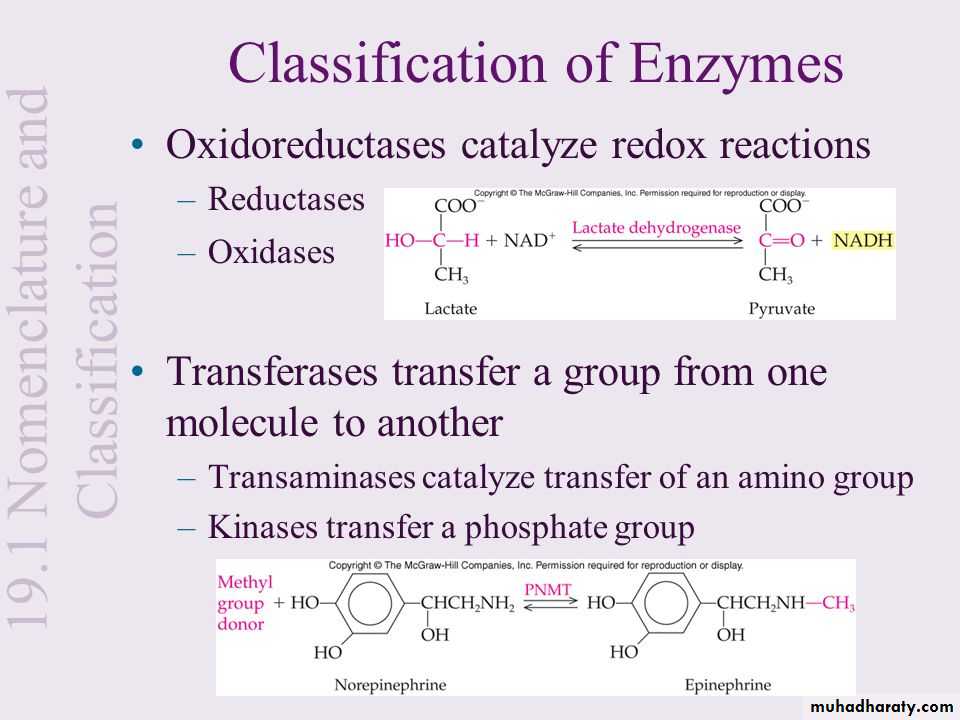
Though enzymes exhibit great degrees of specificity, cofactors may serve many apoenzymes. For example, nicotinamide adenine dinucleotide (NAD) is a coenzyme for a great number of dehydrogenase reactions in which it acts as a hydrogen acceptor. Among them are the alcohol dehydrogenase, malate dehydrogenase and lactate dehydrogenase reactions.
.